Research Group: Cardiovascular Epigenetics (Group Leader: Prof. Dr. med. Francesco Paneni)
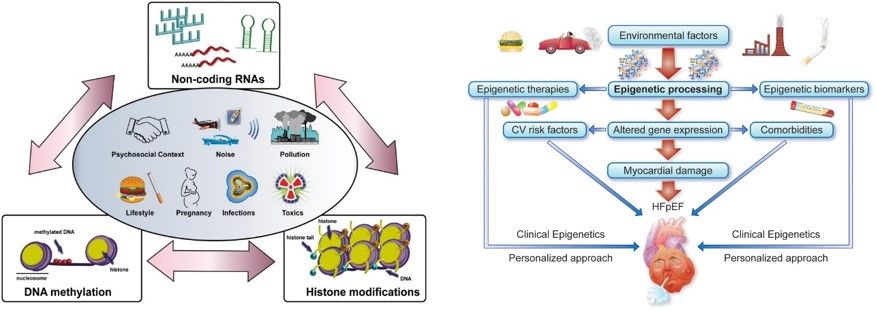
Environmental factors are potent drivers of altered phenotypes and disease states. Exposure to different stimuli may indeed favour detrimental changes eliciting pathological processes in different organs, thus precipitating a cluster of comorbidities such as obesity, diabetes, aging and cardiovascular disease (CVD, Fig. 1).
These conditions often occur simultaneously and significantly aggravate human health by affecting quality of life as well as lifespan. Epigenetic modifications – defined as heritable changes in gene activity that do not affect DNA sequence – may significantly derail transcriptional programs implicated in oxidative stress, inflammation, senescence, defective stem cell functionality, and metabolic alterations, thus fostering maladaptive pathways and premature CVD. Epigenetic signatures may be classified into three main categories: (1) DNA methylation, (2) posttranslational histone modifications, and (3) RNA-based mechanisms including microRNAs and long non-coding RNAs. The complex interplay between these epigenetic signals may provide a molecular framework through which the environment can interact with the genome to alter gene expression and thereby influence cardiovascular homeostasis.
Figure 1. Environmental factors and epigenetics. Over time, an array of environmental factors significantly contributes to build our individual epigenetic background that includes DNA methylation changes, post-translational histone modifications and altered expression of non-coding RNAs. Epigenetic processing takes center stage in several cardiovascular disease, namely cardiac remodeling and heart failure with preserved ejection fraction (HFpEF). Adapted from Costantino…Paneni, Eur Heart J 2019 (left panel) and Hadmani..Paneni, Eur Heart J 2021 (right panel).
We and others have previously shown that epigenetic signatures may be reversible, thus offering exciting opportunities to alter the trajectory of age and diabetes-related CVD. Indeed, plastic epigenetic changes are amenable to pharmacological intervention. Several specific compounds (e.g. BET protein inhibitors) that target enzymes responsible for epigenetic changes [i.e. histone deacetylase (HDAC) and histone acetyltransferase (HATs) inhibitors] have been developed and are in the clinic or in clinical trials to be tested for several age-related and CV diseases (Fig. 2).
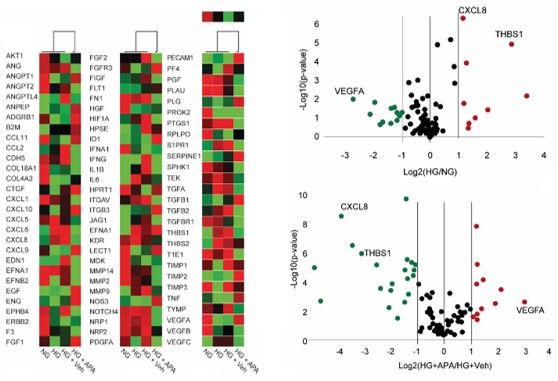
Figure 2. BET protein inhibition resets the endothelial transcriptome. Left panel: Heat map showing differential expression of senescence and inflammatory genes in NG- and HG-treated HAECs, in the presence of RVX-208 (APA, 20 uM) or vehicle (DMSO); n=4/group. Right panel: Volcano plot shows differentially expressed genes in HAECs exposed to NG versus HG. Scatter and volcano plots showing fold changes (log2 values) for inflammaging-related genes in HG-treated HAECs, in the presence of RVX-208 (APA) or vehicle. From Mohammed et al. Antiox. Redox Signalling 2022.
Ongoing research lines
- Understand the role of chromatin remodelling in heart failure with preserved ejection fraction (HFpEF)
- Investigate the function of BET proteins and their therapeutic modulation in cardiometabolic states and age-related diseases
- Decipher the mechanisms underpinning epigenetic inheritance
- Unveil new epigenetic biomarkers of cardiovascular disease
Research Team
Shafeeq Mohammed (Research Assistant)
Era Gorica, PhD (Research Assistant)
Martin Geiger (Research Assistant)
Valentina Delfine (Research Assistant)
Alessia Mongelli (PhD Student)
Funding
Swiss National Science Foundation
Swiss Heart Foundation
Hochschulmedizin Zürich
EU Framework Programme
Novartis Foundation for Biomedical Research
Gebauer Stiftung
Stiftung für wissenschaftliche Forschung
Olga Mayenfisch Foundation
Swiss Life Foundation
Kurt und Senta-Hermann Stiftung
EMDO Stiftung
Schweizerische Diabetes-Stiftung
- Mengozzi A, Costantino S, Paneni F, Duranti E, Nannipieri M, Mancini R, Lai M, La Rocca V, Puxeddu I, Antonioli L, Fornai M, Ghionzoli M, Georgiopoulos G, Ippolito C, Bernardini N, Ruschitzka F, Pugliese NR, Taddei S, Virdis A, Masi S. Targeting SIRT1 Rescues Age- and Obesity-Induced Microvascular Dysfunction in Ex Vivo Human Vessels.
Circ Res. 2022 Aug 15:101161CIRCRESAHA122320888. doi: 10.1161/CIRCRESAHA.122.320888. - Ambrosini S, Montecucco F, Kolijn D, Pedicino D, Akhmedov A, MohammedSA, Herwig M, Gorica E, Lujza Szabó P, Weber L, Russo G, Vinci R, MatterCM, Liuzzo G, Brown PJ, Rossi FMV, Camici GG, Sciarretta S, Paolo BeltramiA, Crea F, Podesser B, Lüscher TF, Kiss A, Ruschitzka F, Hamdani N,Costantino S, Paneni F. Methylation of the Hippo effector YAP by themethyltransferase SETD7 drives myocardial ischemic injury: a translational study.
Cardiovasc Res. 2022. doi: 10.1093/cvr/cvac102. PMID: 35709329 - Mohammed SA, Albiero M, Ambrosini S, Gorica E, Karsai G, Caravaggi CM, Masi S, Camici GG, Wenzl F, Calderone V, Madeddu P, Sciarretta S, Matter CM, Spinetti G, Luscher TF, Ruschitzka F, Costantino S, Fadini GP, Paneni F. The BET Protein Inhibitor Apabetalone Rescues Diabetes-induced Impairment of Angiogenic Response by Epigenetic Regulation of Thrombospondin-1.
Antioxid Redox Signal. 2021. DOI:10.1089/ars.2021.0127. PMID: 34913726 - Hamdani N, Costantino S, Mügge A, Lebeche D, Tschöpe C, Thum T, Paneni F. Leveraging clinical epigenetics in heart failure with preserved ejection fraction: a call for individualized therapies.
Eur Heart J. 2021; 42(20):1940-1958. PMID: 33948637 - Costantino S, Akhmedov A, Melina G, Mohammed SA, Othman A, Ambrosini S, Wijnen WJ, Sada L, Ciavarella GM, Liberale L, Tanner FC, Matter CM, Hornemann T, Volpe M, Mechta-Grigoriou F, Camici GG, Sinatra R, Lüscher TF, Paneni F. Obesity-induced activation of JunD promotes myocardial lipid accumulation and metabolic cardiomyopathy.
Eur Heart J. 2019; 40:997-1008. PMID: 30629164. - Costantino S*, Paneni F*, Virdis A, Hussain S, Mohammed SA, Capretti G, Akhmedov A, Dalgaard K, Chiandotto S, Pospisilik JA, Jenuwein T, Giorgio M, Volpe M, Taddei S, Lüscher TF, Cosentino F. Interplay among H3K9-editing enzymes SUV39H1, JMJD2C and SRC-1 drives p66Shc transcription and vascular oxidative stress in obesity. (*co-first authors).
Eur Heart J. 2019; 40:383-391. PMID: 29077881 - Costantino S, Libby P, Kishore R, Tardif JC, El-Osta A, Paneni F. Epigenetics and Precision Medicine in Cardiovascular Patients: From Basic Concepts to the Clinical Arena.
Eur Heart J. 2018; 39:4150-4158. PMID: 29069341