Research Group Prof. Dr. Reto Schüpbach
Our group is working on the protease-activated receptors (PARs), which play a major role in orchestrating the interactions between coagulation and inflammation. We aim to gain a better understanding of the molecular mechanisms involved in the regulation of PAR activation, which may lead to novel therapeutic options for treating PAR-driven inflammatory and malignant diseases. PAR1, PAR3 and PAR4 are major regulators of platelet activation and vascular barrier function while PAR2 plays a major role in the regulation of extravascular inflammation and cancer.
Proteolytic activation of protease-activated receptors (PARs)
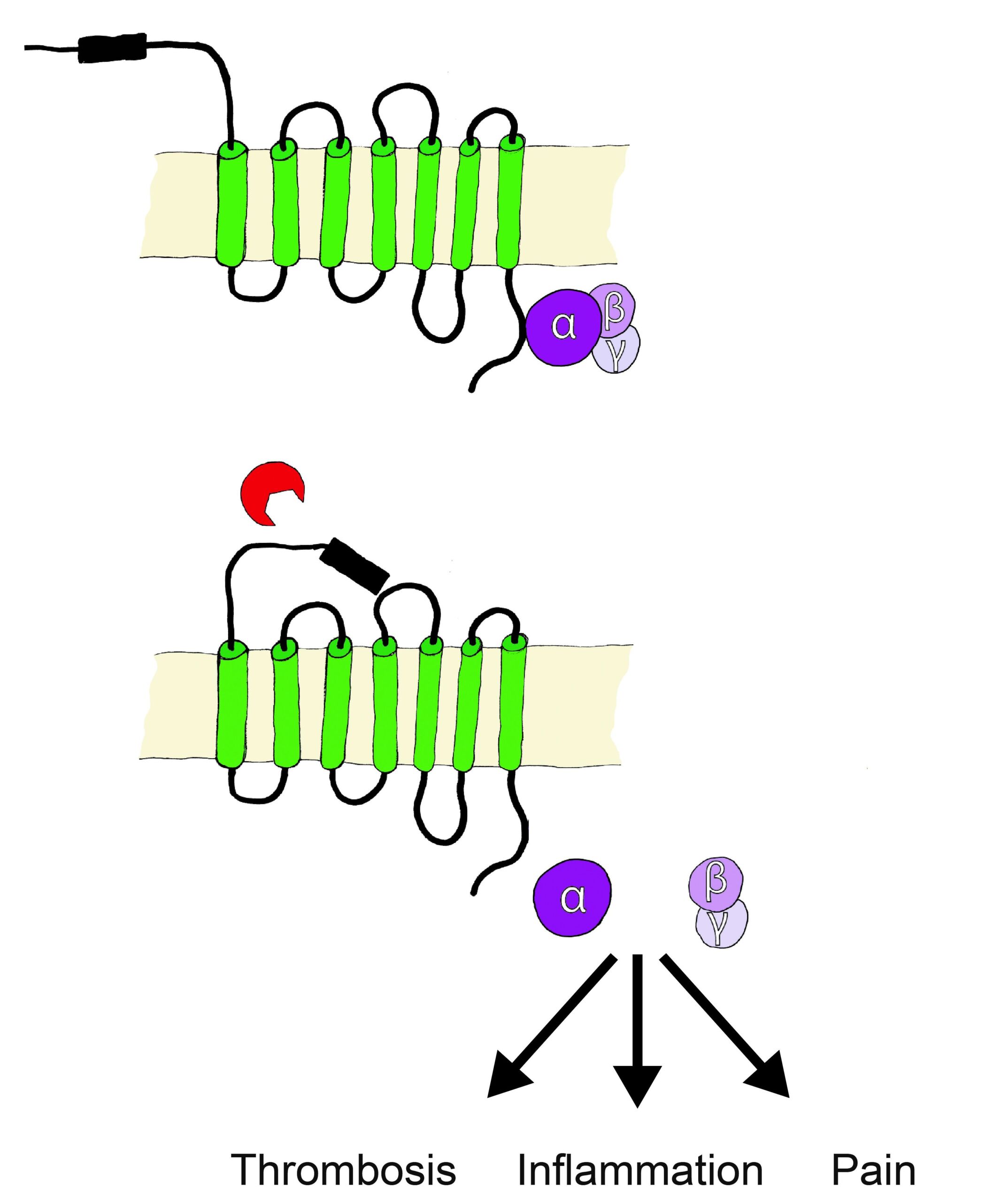
PARs belong to the receptor family of seven transmembrane G-protein coupled receptors (GPCR), however are unique in their lack of physiologically soluble ligands. Proteases, soluble or cell membrane associated (bound to co-receptors or specific membrane compartments) cleave off specific N-terminal sequences of PARs, resulting in the exposure of new N-terminal sequences that serve as tethered activation ligands. The tethered ligands bind to a conserved region on PARs extracellular loop 2, this initiates conformational changes and alters affinity towards intracellular G proteins. The cleavage derived tethered ligands alternatively lead to the transactivation of receptors such as co-localized PARs, ion channels, and toll-like receptors.
PARs are specifically cleaved and irreversibly activated by various endogenous as well as exogenous proteases originating from bacteria, amoeba, plants, fungi, insects or reptiles.
Heuberger and Schüpbach Thromb J. 2019
